A post by Cecina Babich Morrow, PhD student on the Compass programme.
Introduction
Sensitivity analysis seeks to understand how much changes in each input affect the output of a model. We want to be able to determine how variation in a model’s output can be attributed to variations in its input. Given the high amount of uncertainty present in most real-world modelling settings, it is crucial to understand the magnitude of this uncertainty’s impact on model results. Knowing how sensitive a model is to a particular parameter can help guide modellers in prioritising what level of precision is needed in estimating that parameter in order to produce valid results. Sensitivity analysis thus serves as a vital tool for modellers in numerous fields, allowing them to assess robustness and to identify key drivers of uncertainty. By systematically analysing the relative amount of influence that each input parameter has on the output, sensitivity analysis reveals which parameters have the greatest impact on the results.
By identifying these critical parameters, stakeholders can prioritize investments in data collection, parameter estimation, and uncertainty reduction. This targeted approach ensures that efforts are concentrated where they will have the most significant impact.
Why use Regional Sensitivity Analysis?
In this blog post, I will focus on one particular sensitivity analysis method that I have been using in my project so far to help understand the sensitivity of an output decision to input parameters which affect that decision. Regional Sensitivity Analysis (RSA) was developed in the field of hydrology, but has widespread applications in environmental modelling, disease modelling, and beyond.
My research focuses on environmental decision-making, so I frequently deal with models that output a decision that can take on one of several discrete values. For example, consider trying to make a decision about what to wear based on the weather. To make our decision, we use three input parameters about the weather: temperature, humidity, and wind speed. Then, our decision model can output one of three decisions: (1) stay home, (2) leave the house with a jacket, (3) leave the house without a jacket. We might then be interested in how sensitive our model is to each of our three weather-related input parameters to understand how much each one contributes to uncertainty in our ultimate decision. In this type of setting, we need to use a sensitivity analysis method that can handle continuous inputs, e.g. temperature, in conjunction with a discrete output, e.g. our decision.
For settings such as these where the inputs of our model are continuous and the outputs are discrete, RSA, also referred to as Monte Carlo filtering, is a potential method of sensitivity analysis [1]. RSA aims to identify which regions of input space corresponding to specific values in the output space [2, 3]. Originally, the method was developed in the field of hydrology for cases where the output variable is binary, or made such by applying a threshold. It has since been extended by splitting the parameter space into more than two groups [3, 4]. RSA is well-suited to sensitivity analysis in the case where the output variable is categorical [5].
RSA is fundamentally a Bayesian approach. First, prior distributions are assigned to the input parameters. The model is then run multiple times, sampling input parameters from these priors, and recording the resulting output values. By analysing the relationship between input uncertainties and output uncertainties, RSA identifies which parameters significantly affect the model’s predictions.
How does RSA work?
We will present the mathematical formalisation of RSA in a setting where we have a discrete output variable  which can take on one of
which can take on one of 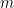 possible output values, and a vector of
possible output values, and a vector of  continuous input variables
continuous input variables ![\mathbf{x} = [x_1, x_2, \ldots, x_d] \mathbf{x} = [x_1, x_2, \ldots, x_d]](https://s0.wp.com/latex.php?latex=%5Cmathbf%7Bx%7D+%3D+%5Bx_1%2C+x_2%2C+%5Cldots%2C+x_d%5D&bg=ffffff&fg=000000&s=0) . We start with prior distributions on the input vector
. We start with prior distributions on the input vector 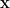 , from which we sample before running the model to calculate the output value for that particular input.
, from which we sample before running the model to calculate the output value for that particular input.
Then, RSA compares the empirical conditional cumulative distribution functions (CDFs)  conditioned on the different output values of
conditioned on the different output values of  . That is, for the
. That is, for the 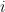 th input parameter, we take the empirical CDF conditioned on the output of the model being the
th input parameter, we take the empirical CDF conditioned on the output of the model being the 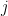 th possible output value. For example, in our weather-based decision model, we would be considering the empirical CDF
th possible output value. For example, in our weather-based decision model, we would be considering the empirical CDF  . We then compare these CDFs
. We then compare these CDFs  for each of the possible
for each of the possible  output values (in our case, each of the possible output decisions). If the conditional CDFs of
output values (in our case, each of the possible output decisions). If the conditional CDFs of  differ greatly in distribution for one or more of the values of
differ greatly in distribution for one or more of the values of  , then we can conclude that our model is sensitive to that particular input parameter. If
, then we can conclude that our model is sensitive to that particular input parameter. If  , then the output is insensitive to
, then the output is insensitive to  on its own. (See the Extensions of RSA section for a discussion of variable interactions.)
on its own. (See the Extensions of RSA section for a discussion of variable interactions.)
The difference between these CDFs can be measured using several possible sensitivity indices. Typically, the Kolmogorov-Smirnov (KS) statistic is applied over all possible values of  , and then some statistic (e.g. mean, median, maximum, etc.) is calculated to summarise the overall sensitivity of
, and then some statistic (e.g. mean, median, maximum, etc.) is calculated to summarise the overall sensitivity of  to
to  :
:
![\text{stat}_{j,k} [KS(x_i)] = \text{stat}_{j,k} \left[\max_{x_i} \left \lvert F_{x_i | y_j} (x_i | y = y_j) - F_{x_i | y_k} (x_i | y = y_k) \right \rvert\right] \text{stat}_{j,k} [KS(x_i)] = \text{stat}_{j,k} \left[\max_{x_i} \left \lvert F_{x_i | y_j} (x_i | y = y_j) - F_{x_i | y_k} (x_i | y = y_k) \right \rvert\right]](https://s0.wp.com/latex.php?latex=%5Ctext%7Bstat%7D_%7Bj%2Ck%7D+%5BKS%28x_i%29%5D+%3D+%5Ctext%7Bstat%7D_%7Bj%2Ck%7D+%5Cleft%5B%5Cmax_%7Bx_i%7D+%5Cleft+%5Clvert+F_%7Bx_i+%7C+y_j%7D+%28x_i+%7C+y+%3D+y_j%29+-+F_%7Bx_i+%7C+y_k%7D+%28x_i+%7C+y+%3D+y_k%29+%5Cright+%5Crvert%5Cright%5D+&bg=ffffff&fg=000000&s=0)
where  and
and  could be mean, median, maximum, etc.
could be mean, median, maximum, etc.
For instance, consider the following situation with an input parameter  , where the output
, where the output  can take on one of three values. We assumed a uniform prior for
can take on one of three values. We assumed a uniform prior for  . The blue, green, and red distributions shown in Fig. 1 below are the empirical conditional CDFs
. The blue, green, and red distributions shown in Fig. 1 below are the empirical conditional CDFs  ,
,  , and
, and  , respectively, giving the probability that
, respectively, giving the probability that  is less than or equal to a given value given that the output result of the model was
is less than or equal to a given value given that the output result of the model was  . The vertical dotted lines are the KS statistic between each of the three pairs of CDFs. Then a statistic, such as the mean, median, or maximum of those three KS values, can be calculated to represent the overall sensitivity of
. The vertical dotted lines are the KS statistic between each of the three pairs of CDFs. Then a statistic, such as the mean, median, or maximum of those three KS values, can be calculated to represent the overall sensitivity of  to the input parameter
to the input parameter  . For example, the mean KS statistic is 0.5505.
. For example, the mean KS statistic is 0.5505.

Figure 1. Visualisation of RSA using a summary statistic of the KS statistic as a sensitivity index. The blue, green, and red distributions are the empirical conditional CDFs  for
for  , and the vertical dotted lines represent the KS statistic between each of the three pairs of CDFs.
, and the vertical dotted lines represent the KS statistic between each of the three pairs of CDFs.
As an alternative to using the KS statistic, we can instead apply a statistic to spread, i.e. the area between the CDFs:
![\text{stat}_{j,k} [\text{spread}(x_i)] = \text{stat}_{j,k} \left[ \int_{-\infty}^\infty \max \left(F_{x_i | y_j} (x_i | y = y_j), F_{x_i | y_k} (x_i | y = y_k)\right) dx_i - \int_{-\infty}^\infty \min \left(F_{x_i | y_j} (x_i | y = y_j), F_{x_i | y_k} (x_i | y = y_k)\right) dx_i \right] \text{stat}_{j,k} [\text{spread}(x_i)] = \text{stat}_{j,k} \left[ \int_{-\infty}^\infty \max \left(F_{x_i | y_j} (x_i | y = y_j), F_{x_i | y_k} (x_i | y = y_k)\right) dx_i - \int_{-\infty}^\infty \min \left(F_{x_i | y_j} (x_i | y = y_j), F_{x_i | y_k} (x_i | y = y_k)\right) dx_i \right]](https://s0.wp.com/latex.php?latex=%5Ctext%7Bstat%7D_%7Bj%2Ck%7D+%5B%5Ctext%7Bspread%7D%28x_i%29%5D+%3D+%5Ctext%7Bstat%7D_%7Bj%2Ck%7D+%5Cleft%5B+%5Cint_%7B-%5Cinfty%7D%5E%5Cinfty+%5Cmax+%5Cleft%28F_%7Bx_i+%7C+y_j%7D+%28x_i+%7C+y+%3D+y_j%29%2C+F_%7Bx_i+%7C+y_k%7D+%28x_i+%7C+y+%3D+y_k%29%5Cright%29+dx_i+-+%5Cint_%7B-%5Cinfty%7D%5E%5Cinfty+%5Cmin+%5Cleft%28F_%7Bx_i+%7C+y_j%7D+%28x_i+%7C+y+%3D+y_j%29%2C+F_%7Bx_i+%7C+y_k%7D+%28x_i+%7C+y+%3D+y_k%29%5Cright%29+dx_i+%5Cright%5D&bg=ffffff&fg=000000&s=0)
where  . In this case, we would be considering the area between each of the three distributions shown in Fig. 1 above and then averaging them (or applying some other summary statistic) as our sensitivity index. For instance, the mean spread between CDFs is 134.09.
. In this case, we would be considering the area between each of the three distributions shown in Fig. 1 above and then averaging them (or applying some other summary statistic) as our sensitivity index. For instance, the mean spread between CDFs is 134.09.
Higher values of either sensitivity index for a given input parameter  suggest that the output is more sensitive to variations in that parameter, i.e. the distributions of input values leading to a given output value are more different from one another. For example, Figure 2 compares the conditional CDFs of
suggest that the output is more sensitive to variations in that parameter, i.e. the distributions of input values leading to a given output value are more different from one another. For example, Figure 2 compares the conditional CDFs of  with that of a different input parameter,
with that of a different input parameter, 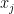 , with a prior of
, with a prior of  . We can see that the CDFs
. We can see that the CDFs  show a high degree of separation, compared to the CDFs
show a high degree of separation, compared to the CDFs  , which do not. This is reflected in the sensitivity indices: for example, the mean KS statistic for
, which do not. This is reflected in the sensitivity indices: for example, the mean KS statistic for 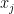 is only 0.1648 and the mean spread is only 2.897. Comparing KS statistics in this manner makes RSA a tool well-suited for ranking, or factor prioritisation, one of the main goals of sensitivity analysis that aims to rank parameters by their contribution to variation in the output [1, 5].
is only 0.1648 and the mean spread is only 2.897. Comparing KS statistics in this manner makes RSA a tool well-suited for ranking, or factor prioritisation, one of the main goals of sensitivity analysis that aims to rank parameters by their contribution to variation in the output [1, 5].

Figure 2. Comparison of sensitivity of a model to two input parameters,  and
and 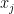 . The blue, green, and red distributions are the empirical conditional CDFs
. The blue, green, and red distributions are the empirical conditional CDFs  and
and  for
for  .
.
Extensions of RSA
One notable limitation of RSA, identified since its inception [2], is its inability to handle parameter interactions. A zero value of the sensitivity index is a necessary condition for insensitivity, but it is not sufficient [2, 5]. Inputs that contribute to variation in the model output only through interactions can have the same univariate conditional CDFs, and thus RSA cannot properly identify their impact on model output. For theoretical examples, see Fig. 1 of [2] and Example 6 of Section 5.2.3 in [1]. In our toy example, we may have a decision model where the output decision is not particularly sensitive to temperature or humidity on their own, but it may be very sensitive to an interaction between these two parameters since their combined effects impact how warm or cool the weather actually feels.
In situations such as these where interactions between input variables may matter more than each variable on its own, RSA can be useful for ranking, but it cannot be used for screening, another goal of sensitivity analysis aiming to identify variables with little to no influence on output variability[1, 5]. To address this limitation, RSA can be augmented with machine learning methods such as random forests and density estimation trees [6]. Spear et al. performed a sensitivity analysis of a dengue epidemic model to demonstrate how these tree-based models can augment RSA [6].
First, the authors performed RSA in its original form, using the KS statistic to examine the difference between the univariate CDFs. Then, they used random forest analysis to classify model runs into the various output values. Then, a measure of variable importance, such as Gini impurity, was used to rank the input parameters in terms of their influence on the model output [6]. Random forest allows for the incorporation of the effects of variable interactions in ranking the importance of each parameter. By comparing the parameter ranking resulting from RSA with that from the random forest, they identified parameters which impacted the output through interaction. Finally, they used density estimation trees to help identify regions of parameter space corresponding to particular output values. Density estimation trees are the analogue of classification and regression trees, instead attempting to estimate the probability density function that gave rise to a particular region of output space [7]. By applying density estimation trees as part of the sensitivity analysis, Spear et al. were able to examine the effects of scale on sensitivity, identifying parameters which may be relatively unimportant when ranking across the entire parameter subspace, but are highly influential in small subspaces.
Further research such as this highlights the benefits of combining multiple sensitivity analysis methods in order to gain a full picture of how model inputs affect uncertainty in the output.
Conclusions
Hopefully this blog has been an informative crash course in regional sensitivity analysis! Note that the visualisations in this post have been created using the SAFEpython toolbox [8]. If you have any questions or comments, please feel free to get in touch at cecina.babichmorrow@bristol.ac.uk.
References
[1] A. Saltelli, Global sensitivity analysis: the primer. Wiley, 2008. [Online]. Available: https://onlinelibrary.wiley.com/doi/book/10.1002/9780470725184
[2] R. Spear and G. Hornberger, “Eutrophication in peel inlet—II. identification of critical uncertainties via generalized sensitivity analysis,” Water Research, vol. 14, no. 1, pp. 43–49, 1980. [Online]. Available: https://www.sciencedirect.com/science/article/pii/0043135480900408
[3] J. Freer, K. Beven, and B. Ambroise, “Bayesian estimation of uncertainty in runoff prediction and the value of data: An application of the GLUE approach,” Water Resources Research, vol. 32, no. 7, pp. 2161–2173, 1996. [Online]. Available: https://onlinelibrary.wiley.com/doi/abs/10.1029/95WR03723
[4] T. Wagener, D. P. Boyle, M. J. Lees, H. S. Wheater, H. V. Gupta, and S. Sorooshian, “A framework for development and application of hydrological models,” Hydrology and Earth System Sciences, vol. 5, no. 1, pp. 13–26, 2001. [Online]. Available: https://hess.copernicus.org/articles/5/13/2001/
[5] F. Pianosi, K. Beven, J. Freer, J. W. Hall, J. Rougier, D. B. Stephenson, and T. Wagener, “Sensitivity analysis of environmental models: A systematic review with practical workflow,” Environmental Modelling & Software, vol. 79, pp. 214–232, 2016. [Online]. Available: https://linkinghub.elsevier.com/retrieve/pii/S1364815216300287
[6] R. C. Spear, Q. Cheng, and S. L. Wu, “An example of augmenting regional sensitivity analysis using machine learning software,” vol. 56, no. 4, p. e2019WR026379. [Online]. Available: https://onlinelibrary.wiley.com/doi/abs/10.1029/2019WR026379
[7] P. Ram and A. G. Gray, “Density estimation trees,” in Proceedings of the 17th ACM SIGKDD international conference on Knowledge discovery and data mining. ACM, pp. 627–635. [Online]. Available: https://dl.acm.org/doi/10.1145/2020408.2020507
[8] F. Pianosi, F. Sarrazin, and T. Wagener, “A Matlab toolbox for global sensitivity analysis,” Environmental Modelling & Software, vol. 70, pp. 80–85, 2015. [Online]. Available: https://linkinghub.elsevier.com/retrieve/pii/S1364815215001188

and
denote the true sensitivity and specificity of Test A,
the true specificity of Test B, and
the prevalence of the condition in the study population. We also introduce
as the covariance of errors between Test A and Test B in the disease-free population.
.

) and specificity (
) of Test A, and hence, the bias. The bias in the naive estimate of specificity is as follows:
), the naive specificity estimate is unbiased. If, however, Tests A and B are conditionally dependent among the disease-free population, then there is a bias which depends on
and
.
), Figure 2 shows
as a function of
, for selected values of
and
.

tends to 1. Bias increases as the accuracy of Test A improves, i.e. when
or
is larger, or with lower prevalence. We can see that when
, the bias is almost negligible. However, bias can be substantial in some scenarios, specifically under low prevalence, even when Test B has high specificity (e.g. over 95%).
).
be the data reported by a `check the negatives’ study evaluating Test A. Then
, where
is the number of participants in the study, and the probabilities
are as specified in Table 1, with
.
could be obtained by fitting this multinomial distribution to the data with vague
priors for the remaining three parameters,
,
, and
.
) is also varied to assess the impact of increasing uncertainty. In Figure 3, we present some result from this simulation study for the correctly centred prior case. These results show that a correctly centred prior consistently eliminates bias under high precision and reduces it under lower precision.




















 The morning started with four ten-minute research talks from Compass students. First was
The morning started with four ten-minute research talks from Compass students. First was  Student Lightning Talks
Student Lightning Talks This year’s conference panel, chaired by Compass CDT Director,
This year’s conference panel, chaired by Compass CDT Director,  The conference concluded with an enlightening special guest lecture by
The conference concluded with an enlightening special guest lecture by  The successful day of talks, poster sessions and networking culminated with Professor Whiteley sharing his thoughts on what we learned throughout the event. He concluded that the future of our field is certain to be exciting and will encompass a huge range of different areas and ideas. This year’s conference embodied this by providing a platform for students, academics, and industry professionals to share new insights from many different sectors, and to form strong relationships to help forge a path to the future of data science.
The successful day of talks, poster sessions and networking culminated with Professor Whiteley sharing his thoughts on what we learned throughout the event. He concluded that the future of our field is certain to be exciting and will encompass a huge range of different areas and ideas. This year’s conference embodied this by providing a platform for students, academics, and industry professionals to share new insights from many different sectors, and to form strong relationships to help forge a path to the future of data science.


